Since the completion of the Human Genome project, one of the key challenges has been correlating genotype with a presented phenotype to identify the potential cause of rare diseases.
Moreover, the rapid advancement of sequencing technology, both in the output of sequencing information and the significant reduction in cost, has created a massive wealth of data resulting in a big data interpretation challenge to understand the genetic causes of disease.
The Deciphering Developmental Disorders (DDD) study was a landmark project, focusing on understanding the genetic basis and improving the diagnosis of patients with developmental disorders. In total, over 13,000 undiagnosed children and adults were recruited during the study through the regional genetics services in the UK and the Republic of Ireland.
-1.png?width=1920&height=1080&name=Deciphering%20Developmental%20Disorders%20(1)-1.png)
In order to learn more about this project as well as advancements in data analysis and interpretation, we hosted a webinar with Professor Matthew Hurles, project lead for the DDD study and Congenica founder to discuss:
- Lessons learnt from the DDD study of post-natal development disorders
- Lessons learnt from the Prenatal Assessment of Genomes and Exomes (PAGE) Study of pre-natal development disorders
- The power of sharing data through the DECIPHER portal
- The future of variant interpretation: large scale functional data
With lots of interest in this webinar and many questions to Professor Matthew Hurles after his presentation, we were unfortunately not able to answer all. However, we have posted these questions to Professor Matthew Hurles and the experts at Congenica, who have kindly provided the answers below.
Question: Are reclassifications being considered for genes where phenotype is clearly very similar yet VUS classification dominant in result given many were classified by DDD 5 years+ ago? (example CACNA1C where knowledge and understanding increased recently) Families arguably still on diagnostic odyssey.
Professor Matthew Hurles: "DDD bioinformatics team reanalyses patient data to identify plausibly pathogenic variants that have not previously been reported to the clinical team caring for the patient, however, once a variant has been reported to the clinical team, they determine whether a historical VUS is reclassified or not. My understanding is that reclassification of variants previously classified as VUS is something that the NHS clinical teams recognise as being important, and is likely to be undertaken as part of ongoing appointments with undiagnosed families. I don’t know whether some centres also reclassify historical VUS outside of these appointments, but I suspect that this might be limited by bandwidth of diagnosticians and there may be differences in practice between different clinical centres."
Question: Has the data in the slide below already been published?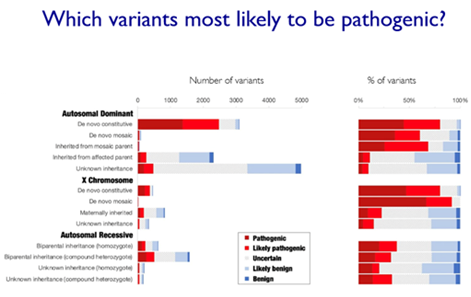
Professor Matthew Hurles: "The manuscript describing this work is under review, but the preprint is available on MedRxiv"
Question: Would it be possible for Congenica to determine the disease prevalence in the UK population?
Congenica: "We are able to collate variants, classifications and HPOs across sites using the Congenica platform, but everything is HPO driven rather than disease driven (e.g. Intellectual disability, hypotelorism, short stature vs Down Syndrome). However, not all patients would get NGS testing (e.g. suspected cystic fibrosis would get a targeted test, not WES or WGS; or hypercholesterolemia would get a blood test, not a DNA test). Potentially a large portion of the more common disorders would be underrepresented, if they are being tested by other methods. Therefore, there might be significant limitations on any conclusions we would be able to draw."
Question: With WGS analysis now only taking a few hours and the reduced cost of $200, what impact will this have on patients with Rare Disease?
Congenica: "The reduced cost of WGS and the implementation of the Congenica clinical decision support platform will allow WGS to become more accessible, enable a faster and more complete genetic diagnosis, and significantly reduce the diagnostic odyssey that rare disease patients and their families currently experience."
Question: What developments and updates can we expect from Congenica in the future?
Congenica: "One of our biggest developments in the coming year is expanding the Congenica decision support platform into oncology. For rare and inherited disease, we continue to develop our platform through a range of updates, including increasing the breadth of interpretable variant types. Our Clinical Interpretation Service (CIS), launched in early 2022, continues to expand and provide interpretation support to more genetic testing laboratories for panels, exomes and genomes".
We would like to thank Professor Matthew Hurles again for a fantastic talk and the excellent discussion. If you would like to watch the recorded webinar, please click the link below. 
Find out more information about Congenica’s automated and AI-powered Clinical Decision Support platform.
-1.png)

.png?width=320&height=192&name=Transforming%20Cancer%20Care%20Using%20Genomic%20Tests%20(2).png)
.png?width=320&height=192&name=Untitled%20design%20(8).png)