Dr Charles Steward
Clinical Domain Lead, Epilepsy, Congenica
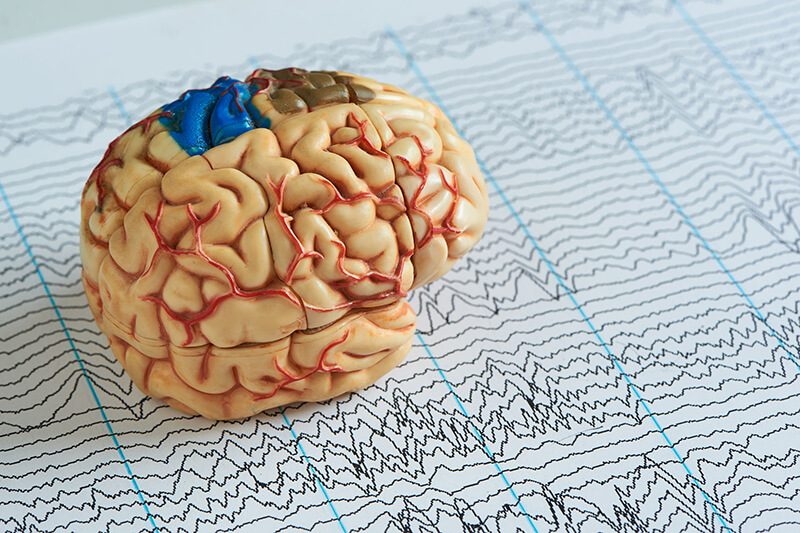
The developmental and epileptic encephalopathies are a group of rare, severe neurodevelopmental disorders, where even after the most thorough sequencing investigations, 60-65% of patients remain without a molecular diagnosis.
The ability to interpret pathogenic variants in patients’ genomes relies upon the completeness of the reference data sets that are used to analyse their genomes.
For example, while we know that the human genome is still not 100% complete, improvements in diagnostic yield can be achieved through improvements in gene annotation. Gene annotation is the process by which a function is assigned to specific sequences in the genome that are
responsible for making proteins.
At the current time, most gene annotations from Ensembl and RefSeq are based on RNA transcript sequences that were produced alongside the initial experimental phase of the Human Genome Project, while the large datasets produced by more recent RNA-Seq and long-read sequencing-based projects remain largely unincorporated.
In our paper, “Re-annotation of 191 Developmental and Epileptic Encephalopathy-associated genes unmasks de novo variants in SCN1A”, we explored the incompleteness of Ensembl transcript models used for exome and genome analysis, as one potential explanation for a lack of current diagnoses for people with epilepsy.
We re-interpreted 191 epilepsy-associated genes using human brain-derived transcriptomic libraries and other data to identify many new exons that were previously missing from the public datasets.
Among the many interesting findings, we discovered three variants in CDKL5 which were previously considered to be intronic but are now shown to lie within a new coding exon.
We used SCN1A as a case study, due to its strong phenotype/genotype correlation with Dravet syndrome, to screen 122 individuals using our new sequences and identified two de novo SCN1A variants in two patients in so called ‘poison exons’.
We also found a previously classified intronic SCN1A Dravet syndrome-associated de novo variant that lies within a new poison exon as well as poison exons in the related genes SCN2A and SCN8A.
Our findings suggest that there are potentially additional causative genetic variants to be identified in epilepsy-related genes.
Furthermore, if patients who currently have no molecular diagnosis are reanalysed using our new exon annotations, we would expect to find some resolvable cases in the gene that was originally suspected by the clinician.
Read the Full Paper
The full paper (DOI: 10.1038/s41525-019-0106-7) is available as an open access paper in npj Genomic Medicine: https://www.nature.com/articles/s41525-019-0106-7
To see how Congenica can help you increase efficiency in the diagnosis of developmental and epileptic disorders


.png?width=320&height=192&name=Untitled%20design%20(8).png)
.png?width=320&height=192&name=Since%202016%2c%20the%20number%20of%20women%20working%20in%20STEM%20fields%20in%20the%20UK%20has%20increased%20by%20216%2c552%2c%20taking%20the%20total%20number%20over%20the%201%20million%20mark%20for%20the%20first%20time.%20Women%20now%20make%20up%2024%25%20of%20the%20STEM%20workforce%20i%20(2).png)
-1.png?width=320&height=192&name=Deciphering%20Developmental%20Disorders%20(1)-1.png)