At the 25th International Conference on Prenatal Diagnosis and Therapy (ISPD), two members of Congenica’s Clinical Interpretation Services team presented posters.
Rand Dubis, Translational Scientist, presented: ‘The Impact of Molecular Diagnosis of Fetal Structural Anomalies Using Exome Sequencing’.
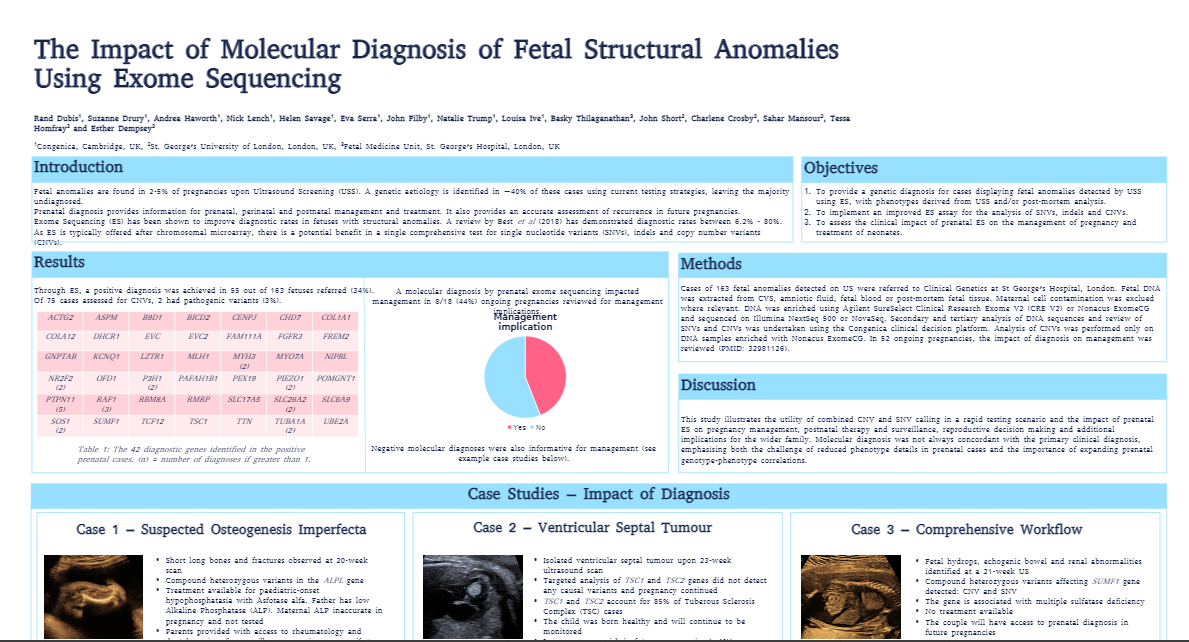
Fetal anomalies are found in 2-5% of pregnancies upon Ultrasound Screening (USS). A genetic aetiology is achieved in ~40% of these cases using current testing strategies, leaving the majority undiagnosed.
This study set out to:
- Provide a genetic diagnosis for cases displaying fetal anomalies detected by USS using Exome Sequencing (ES), with phenotypes derived from USS and/or post-mortem analysis.
- Implement an improved exome sequencing assay for the analysis of SNVs, indels and CNVs.
- Assess the clinical impact of prenatal ES on the management of pregnancy and treatment of neonates.
This poster illustrates the utility of combined CNV and SNV calling in a rapid testing scenario and the impact of prenatal ES on pregnancy management, postnatal therapy and surveillance, reproductive decision making and additional implications for the wider family.
Molecular diagnosis was not always concordant with the primary clinical diagnosis, emphasizing both the challenge of reduced phenotype details in prenatal cases and the importance of expanding prenatal genotype-phenotype correlations.
Suzanne Drury, Lead Translational Scientist, Prenatal Genomics & Personalized Health, presented: 'Non-invasive prenatal solutions for multiple single gene disorders in a single test (INGENIOUS - Improved Next Generation Sequencing for Ultrasound Abnormalities)'.
Fetal anomalies are detected in ~3% of pregnancies and are responsible for ~20% of perinatal deaths. Approximately two-thirds of single gene causes of fetal anomalies are de novo mutations (DNMs). Advances in genomic technology have led to rapid adoption of non-invasive prenatal testing for aneuploidy, but tools to support non-invasive diagnosis of single gene disorders are limited.
Our objective was to develop a comprehensive assay and analytical pipeline to pair with the Congenica clinical decision support platform, to enable non-invasive detection of de novo mutations associated with fetal anomalies.
Our non-invasive approach correctly identified all pathogenic de novo variants, in RAF1, SOS1, FGFR3, COL1A1 and NRAS (x2). Assay performance and analytical annotation and interpretation of genetic variants are crucial for the accurate and rapid reporting of diagnosis in the prenatal period.