 Suzanne Drury, PhD; Clinical Genomics and Personalized Medicine Specialist, Congenica
Suzanne Drury, PhD; Clinical Genomics and Personalized Medicine Specialist, Congenica
Implementation of genomics is providing answers in clinically urgent prenatal settings, impacting medical management, informing therapeutic treatment and enabling couples to plan for the future.
In this post I review prenatal diagnosis in the era of genomic medicine.
Prenatal diagnosis in the era of genomic medicine
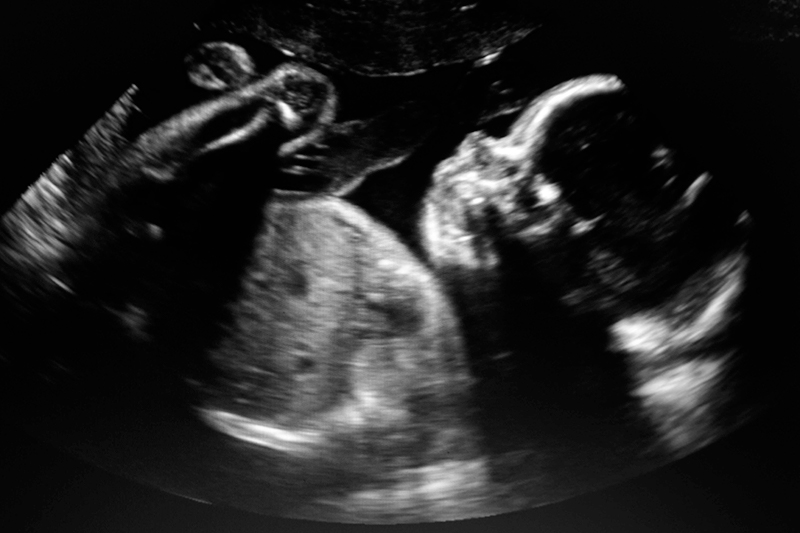
Genetic disorders are a leading cause of morbidity and premature death in the prenatal and neonatal period [1]. Approximately 3% of fetuses have a structural abnormality detected via ultrasound scans, the majority of which remain undiagnosed after routine genetic testing.
However, advances in genomics using exome sequencing and decision support technologies for rapid analysis are revolutionizing prenatal diagnosis and practice, helping to improve diagnostic rates and providing families with answers.
Diagnosis of prenatal ultrasound detected abnormalities
Resolving differential diagnosis in a clinically meaningful timeframe can have a significant impact on the pregnancy. Features on ultrasound which have an unclear prognosis can be resolved by genetic diagnosis, and can provide reassurance, enable tailored counselling, treatment and pregnancy management.
Karyotype, qfPCR, chromosomal microarray (CMA) and non-invasive prenatal testing are the methods which have most commonly been used for prenatal diagnosis of chromosomal anomalies.
However, prior to the availability of exome sequencing, methods for identifying single gene disorders that could be offered in the course of pregnancy was limited to a handful of genes with clear genotype-phenotype association. Mutations in FGFR3, for example are associated with achondroplasia and thanataphoric dysplasia (TD), the most common non-lethal and lethal skeletal dysplasias respectively. Both these conditions are clinically indicated by short limbs on ultrasound, with TD further categorized radiologically based on femur and skull shape.
In the absence of a targeted gene approach, sequential single-gene testing by traditional Sanger sequencing methods could not yield a result in a clinically meaningful timeframe.
Hundreds of single-gene disorders can present prenatally and the majority of the approximately 4,000 known monogenic rare diseases present in the first five years of life. Monogenic disorders account for a proportion of chromosomally normal cases. Whilst exome sequencing is now commonplace in the molecular diagnosis of postnatal conditions, prenatal exome sequencing (pES) is gradually being adopted in the clinic, with prospective cohort studies of exome sequencing for fetal anomalies showing an overall diagnostic yield of 8.5-10% with skeletal and multisystem abnormalities in particular having a higher diagnostic rate (15-24% and 15-19% respectively). Approximately 60% of monogenic causes have been shown to be de novo in unselected cases of fetal anomaly [2,3].
Challenges of Prenatal Genomics
The implementation of extended prenatal genetic testing has so far been based on genetic material obtained using invasive sample collection methods. These methods typically involve collecting amniotic fluid or chorionic villus samples. Whilst the risk of these procedures of causing a miscarriage is small (<0.1-0.3% [4]), many couples choose not to undergo invasive testing.
However, the widespread uptake of non-invasive prenatal testing (NIPT) for common aneuploidy (the presence of an abnormal number of chromosomes in a cell), is leading to increased acceptance of prenatal testing using maternal blood samples. This is also leading to increased acceptance for prenatal testing of serious childhood onset conditions caused by genetic disorders [5] and it is anticipated that development and uptake in this area will continue to grow [6,7].
Whilst, chromosomal microarray is still typically performed prior to exome sequencing, for clinical presentations strongly suggestive of a single gene disorder, performing in parallel would reduce time to diagnosis.
Combined methods for detecting both single nucleotide and copy number variants from a single test are desirable to maximize diagnostic yield, reduce time and cost.
A lack of prenatal data
One of the key challenges of prenatal diagnosis is that phenotype information is limited and non-specific as the fetus is still developing. This means most genotype-phenotype databases have been created using postnatal cases.
Diagnostic tests, such as biochemical assays, which would be used to guide and support diagnosis in the postnatal setting, are difficult or not possible at all. In the fetus, features which can be detected are limited to those on ultrasound and magnetic resonance imaging (MRI) and requires the expertise of a high-quality fetal medicine unit.
Improving prenatal outcomes
Abnormalities which can be detected on fetal ultrasound fall into the following broad categories: skeletal, brain, cardiac, spinal, hydrops, increased nuchal translucency, facial, renal, chest, abdominal and multisystem[2]. Involvement of the clinical genetics team at the time of fetal anomaly scanning in cases of suspected genetic conditions is important in refining clinical diagnosis and obtaining relevant family history. By including clinical geneticists at the referral stage, overall diagnostic rates can increase to over 40% [8].
If a suspected clinical diagnosis is made, this can prompt review of a specific set of genes. However, in these early presentation scenarios the molecular diagnosis is not always the primary clinical diagnosis [8]. Using phenotype-driven gene panels, stratifying analysis initially with genes associated with the queried clinical diagnosis, then expanding to phenotype relevant gene panels if negative is a powerful approach for both reducing the time to diagnosis maintaining diagnostic yield and reducing incidental findings.
Impact of a molecular diagnosis in utero
Prenatal diagnosis is valued by parents, and can aid planning for optimal perinatal management, including delivery and access to immediate neonatal care and can help parents prepare for a future with an affected child.
In the UK, ~1/7 babies are admitted to a neonatal unit each year and studies have shown that around 20% of critically ill children have a known monogenic disease [1]; identifying the genetic cause of disease before birth can avoid unnecessary and often invasive investigative procedures and ensure that optimal care is provisioned at the right time.
Parallel advances in gene editing approaches is raising the potential of in utero gene therapy, where genetic disease could be treated before the onset of irreversible pathology and rescue diseases which are perinatal lethal or cause severe morbidity [9].
Reproductive decision making is also influenced by molecular diagnosis; many couples desire a diagnosis to understand the risk of recurrence in subsequent pregnancies. With a molecular diagnosis, preimplantation or prenatal genetic diagnosis can be an option for informing future pregnancies.
Considerations for counselling
It should be appreciated that not all families choose to undertake genetic testing. It has been discussed how ensuring informed consent in the rapid genomic testing scenario is challenging; there is little time to reflect before making a decision and parents are likely to be under considerable distress [10].
Realistic expectations should be set and the potential impact of both a diagnosis and no diagnosis should be discussed, including uncertain results [11]. Other considerations include whether secondary and incidental findings should be returned, either in regards to the child or to the parent; for example should findings in the ACMG59 genes be reported to the parents regarding their health, and when, even if the primary indication for testing was a fetal anomaly [12].
Accelerating prenatal genomic analysis
The time to diagnosis is critical in the prenatal setting; the number of genes which can be associated with fetally relevant conditions and continued expansion of understanding of causative genes can cause a challenge for interpretation to even the experienced diagnostician reviewing prenatal genomic data. Trio testing (of both parents and fetus) is recommended, as this reduces time taken for variant interpretation and assists in classification of pathogenicity.
To address some of these challenges, we have developed Congenica Prenatal™, a pre-configured application within the Congenica clinical decision support platform, to provide accurate and rapid identification of de novo variants in genes associated with autosomal dominant and x-linked fetal anomalies.
Annotations within Congenica Prenatal signpost users to relevant literature describing fetal presentations associated with genes in expert curated gene panels. Curated variant lists further highlight previously identified pathogenic variants in relevant genes and templated reporting streamlines analytical workflows.
Congenica Prenatal stratifies prenatal analysis to enable identification of the molecular cause of fetal anomalies, faster, helping to reduce interpretation times and enable a rapid response.
References
1 Wojcik MH, Schwartz TS, Thiele KE, et al. Infant mortality: the contribution of genetic disorders. J Perinatol Published Online First: 8 August 2019. doi:10.1038/s41372-019-0451-5
2 Lord J, McMullan DJ, Eberhardt RY, et al. Prenatal exome sequencing analysis in fetal structural anomalies detected by ultrasonography (PAGE): a cohort study. Lancet 2019;393:747–57. doi:10.1016/S0140-6736(18)31940-8
3 Petrovski S, Aggarwal V, Giordano JL, et al. Whole-exome sequencing in the evaluation of fetal structural anomalies: a prospective cohort study. Lancet Published Online First: 31 January 2019. doi:10.1016/S0140-6736(18)32042-7
4 American College of Obstetricians and Gynecologists’ Committee on Practice Bulletins—Obstetrics, Committee on Genetics, Society for Maternal–Fetal Medicine. Practice Bulletin No. 162. Obstet Gynecol 2016;127:e108–22. doi:10.1097/AOG.0000000000001405
5 Hill M, Compton C, Karunaratna M, et al. Client Views and Attitudes to Non-Invasive Prenatal Diagnosis for Sickle Cell Disease, Thalassaemia and Cystic Fibrosis. J Genet Couns 2014;23:1012–21. doi:10.1007/s10897-014-9725-4
6 Sullivan HK, Bayefsky M, Wakim PG, et al. Noninvasive Prenatal Whole Genome Sequencing. Obstet Gynecol 2019;:1. doi:10.1097/AOG.0000000000003121
7 Drury S, Hill M, Chitty LS. Cell-Free Fetal DNA Testing for Prenatal Diagnosis. In: Advances in Clinical Chemistry. Academic Press Inc. 2016. 1–35. doi:10.1016/bs.acc.2016.05.004
8 Drury S, Reed L, Ramachandran V, et al. Diagnosis of fetal structural abnormalities using whole exome sequencing: a single centre study. 2019.https://www.abstractsonline.com/pp8/#!/7874/presentation/732
9 Peranteau WH, Flake AW. The Future of In Utero Gene Therapy. Mol Diagn Ther 2020;24. doi:10.1007/s40291-020-00445-y
10 Gyngell C, Newson AJ, Wilkinson D, et al. Rapid Challenges: Ethics and Genomic Neonatal Intensive Care. Pediatrics 2019;143:S14–21. doi:10.1542/peds.2018-1099D
11 Harding E, Hammond J, Chitty LS, et al. Couples experiences of receiving uncertain results following prenatal microarray or exome sequencing: A mixed‐methods systematic review. Prenat Diagn 2020;:pd.5729. doi:10.1002/pd.5729
12 Amor DJ, Chitty LS, Van den Veyver IB. Current controversies in prenatal diagnosis 2: The 59 genes ACMG recommends reporting as secondary findings when sequencing postnatally should be reported when detected on fetal (and parental) sequencing. Prenat Diagn Published Online First: 24 February 2020. doi:10.1002/pd.5670

.png?width=320&height=192&name=Untitled%20design%20(8).png)
.png?width=320&height=192&name=Since%202016%2c%20the%20number%20of%20women%20working%20in%20STEM%20fields%20in%20the%20UK%20has%20increased%20by%20216%2c552%2c%20taking%20the%20total%20number%20over%20the%201%20million%20mark%20for%20the%20first%20time.%20Women%20now%20make%20up%2024%25%20of%20the%20STEM%20workforce%20i%20(2).png)
-1.png?width=320&height=192&name=Deciphering%20Developmental%20Disorders%20(1)-1.png)