At the Curating the Clinical Genome event in May 2021, Suzanne Drury, Lead Transitional Scientist, Prenatal Genomics and Personalized Health at Congenica, presented a poster that detailed non-invasive prenatal solutions for multiple single gene disorders in a single test - Improved Next Generation Sequencing for Ultrasound Abnormalities (INGENIOUS).
Introduction
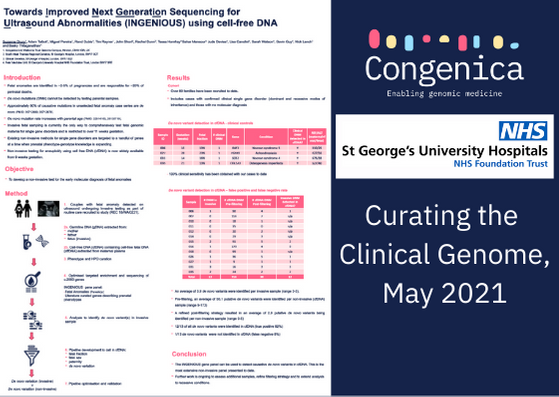
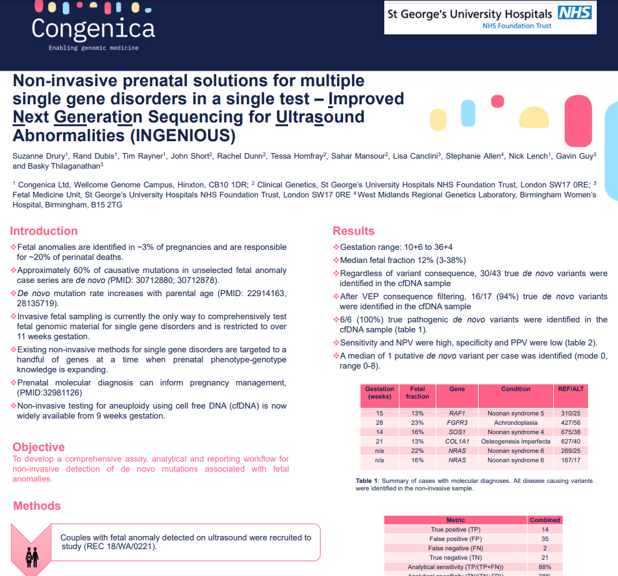
Fetal anomalies are detected in ~3% of pregnancies and are responsible for ~20% of perinatal deaths. Approximately two-thirds of single gene causes of fetal anomalies are de novo mutations (DNMs). Advances in genomic technology have led to rapid adoption of non-invasive prenatal testing for aneuploidy, but tools to support non-invasive diagnosis of single gene disorders are limited.
Our objective was to develop a comprehensive assay and analytical pipeline to pair with the Congenica clinical decision support platform, to enable non-invasive detection of de novo mutations associated with fetal anomalies.
The INGENIOUS gene panel detects causative de novo variants in cfDNA
Cases from 73 quads with gestation range 10+6 to 36+4 were assessed and had a median fetal fraction of 12% (range 3-38%). In total 17 de novo variants were identified and 16/17 (94%) were detected in the non-invasive sample. Six of these variants were confirmed to be pathogenic and 100% (6/6) were correctly identified in cfDNA. A median of 1 putative de novo variant per case was identified (mode 0, range 0-8).
Other recent posters presented by Congenica include:
Automated variant classification

.png?width=320&height=192&name=Untitled%20design%20(8).png)
.png?width=320&height=192&name=Since%202016%2c%20the%20number%20of%20women%20working%20in%20STEM%20fields%20in%20the%20UK%20has%20increased%20by%20216%2c552%2c%20taking%20the%20total%20number%20over%20the%201%20million%20mark%20for%20the%20first%20time.%20Women%20now%20make%20up%2024%25%20of%20the%20STEM%20workforce%20i%20(2).png)
-1.png?width=320&height=192&name=Deciphering%20Developmental%20Disorders%20(1)-1.png)